Inosine (I) in RNA Structure and Function: A Versatile Nucleoside
What is Inosine?
Inosine is a nucleoside that occurs naturally in transfer RNAs (tRNAs) and is essential for proper RNA translation. It is commonly found at the wobble position of anticodons, allowing for greater flexibility in base pairing. Inosine is also formed by the deamination of adenosine, a process catalyzed by enzymes called ADARs (adenosine deaminases acting on RNA).
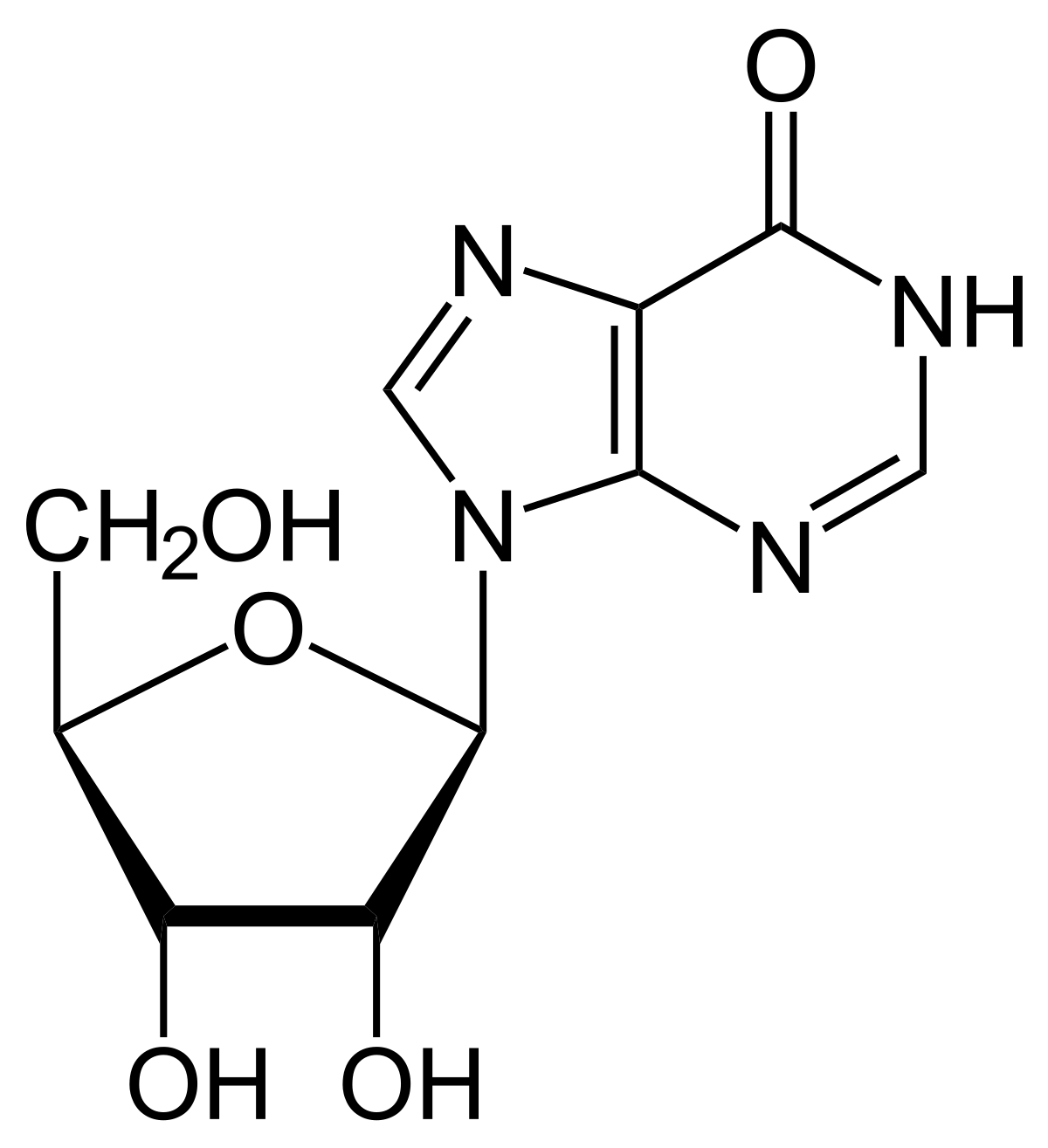
Inosine in tRNA and the Wobble Hypothesis
The wobble hypothesis, proposed by Francis Crick in 1966, suggests that the pairing between the third base of a codon and the corresponding base of an anticodon is less stringent than the standard Watson-Crick base pairing. This flexibility is often mediated by inosine at the wobble position (first position) of the anticodon. Inosine can pair with adenine, cytosine, or uracil, thus expanding the decoding capacity of a single tRNA.
Inosine Base Pairing
Inosine forms two hydrogen bonds with adenine, similar to the Watson-Crick base pairing between adenine and uracil. However, inosine can also form wobble base pairs with cytosine and uracil, although these interactions are less stable. The versatile base pairing properties of inosine allow it to decode multiple codons, reducing the number of required tRNA species.
A-to-I RNA Editing
Inosine can also be introduced into RNA through a process called A-to-I RNA editing. In this process, adenosine deaminases acting on RNA (ADARs) convert adenosine (A) to inosine (I) by hydrolytic deamination. Since inosine is interpreted as guanosine (G) by the cellular machinery, A-to-I editing can alter codons, splicing sites, and miRNA binding sites, leading to changes in protein sequence, alternative splicing, and gene regulation.
ADAR Enzymes
ADARs are a family of enzymes that catalyze the hydrolytic deamination of adenosine to inosine in double-stranded RNA (dsRNA) substrates. Mammals have three ADAR genes: ADAR1, ADAR2, and ADAR3. ADAR1 and ADAR2 are catalytically active and responsible for the majority of A-to-I editing events. These enzymes play crucial roles in regulating gene expression and have been implicated in various biological processes, including development, immunity, and neurological functions.
Inosine in Biotechnology and Therapeutics
The unique properties of inosine have been exploited in various biotechnological and therapeutic applications:
Universal Base in Primers and Probes
Inosine can be incorporated into primers and probes as a universal base due to its ability to pair with multiple nucleotides. This property is particularly useful in applications such as degenerate PCR, where a single primer containing inosine can amplify a variety of related sequences, reducing the need for complex primer mixtures.
Inosine Prodrugs
Inosine can be used as a prodrug to deliver purine nucleosides, such as adenosine and guanosine, for therapeutic purposes. For example, inosine pranobex, a synthetic derivative of inosine, has been used as an immunomodulatory and antiviral agent. It is thought to enhance the body's natural defense mechanisms and has been studied for the treatment of various viral infections and immune disorders.
Challenges and Future Perspectives
Despite the significant advances in understanding the roles of inosine in RNA biology, several challenges remain. One of the main challenges is elucidating the precise mechanisms by which A-to-I editing influences gene expression and cellular functions. The identification of novel ADAR substrates and the development of high-throughput screening methods will be crucial in unraveling the full extent of A-to-I editing in the transcriptome.
Another challenge lies in the potential off-target effects of using inosine-containing primers and probes in biotechnological applications. The development of more specific and sensitive tools that can distinguish between inosine and other nucleotides will be essential for accurate and reliable results.
Future research on inosine will likely focus on its role in disease pathogenesis and its potential as a therapeutic target. A-to-I editing has been implicated in various neurological disorders, cancers, and viral infections. Understanding the mechanisms by which altered A-to-I editing contributes to these diseases may open new avenues for diagnosis and treatment. Additionally, the development of novel inosine-based prodrugs and the optimization of existing ones could lead to improved therapies for a range of conditions.
As the field of epitranscriptomics continues to evolve, the study of inosine and other RNA modifications will undoubtedly expand. The integration of high-throughput sequencing technologies, bioinformatics tools, and structural biology approaches will provide a more comprehensive understanding of the complex interplay between RNA modifications, structure, and function. This knowledge will not only deepen our understanding of fundamental biological processes but also pave the way for novel biotechnological and therapeutic applications.
Further Reading
Frontiers in Pharmacology, Inosine: A bioactive metabolite with multimodal actions in human diseases
