Epitranscriptomics: The Emerging Science of RNA Modifications
What is Epitranscriptomics?
Epitranscriptomics is an emerging field that focuses on the study of chemical modifications in RNA molecules and their impact on gene expression, cellular processes, and disease states. These modifications, collectively known as the epitranscriptome, add an additional layer of complexity to the regulation of gene expression beyond the DNA sequence and transcription.
Key Concepts in Epitranscriptomics
Epitranscriptomics revolves around several key concepts:
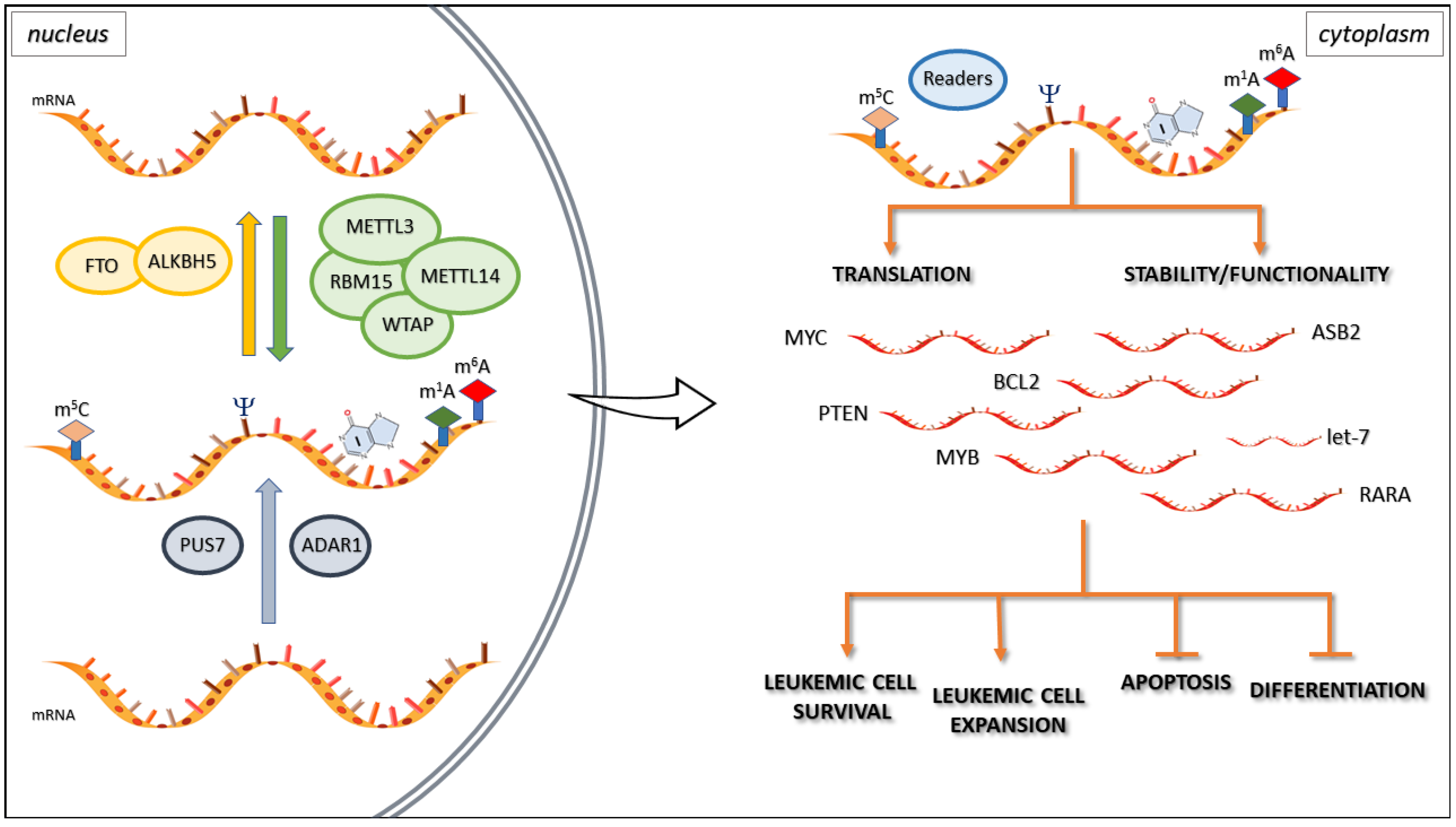
- RNA Modifications: Over 170 different chemical modifications have been identified in various types of RNA, including messenger RNA (mRNA), transfer RNA (tRNA), and ribosomal RNA (rRNA). The most prevalent and well-studied modification is N6-methyladenosine (m6A), but others such as pseudouridine (Ψ), inosine (I), and 5-methylcytosine (m5C) also play important roles.
- Dynamic and Reversible: Many RNA modifications are dynamic and reversible, with specific enzymes responsible for adding (writers), removing (erasers), and recognizing (readers) these modifications. This dynamic nature allows for rapid changes in gene expression in response to cellular signals or environmental cues.
- Functional Consequences: RNA modifications can impact various aspects of RNA biology, including stability, translation efficiency, splicing, localization, and interactions with proteins. These functional consequences ultimately influence gene expression and cellular processes, such as cell differentiation, stress response, and development.
Biological Significance of RNA Modifications
RNA modifications play crucial roles in various biological processes and have been implicated in human health and disease:
Cellular Differentiation and Development
Epitranscriptomic changes have been observed during cellular differentiation and development, suggesting a role in regulating cell fate decisions. For example, m6A modifications in mRNA have been shown to control embryonic stem cell differentiation and neuronal development.
Stress Response and Environmental Adaptation
RNA modifications can be dynamically regulated in response to various cellular stresses, such as heat shock, oxidative stress, and nutrient deprivation. These modifications help cells adapt to changing environments by fine-tuning gene expression and modulating stress response pathways.
Disease Pathogenesis
Dysregulation of RNA modifications has been linked to various human diseases, including cancer, neurological disorders, and metabolic diseases. For instance, altered m6A levels have been observed in several types of cancer, and mutations in m6A-related genes have been associated with neurological disorders such as intellectual disability and autism spectrum disorder.
Epitranscriptomics Methods and Technologies
Advances in sequencing technologies and biochemical methods have enabled the genome-wide mapping and functional characterization of RNA modifications. Some key techniques used in epitranscriptomics research include:
High-Throughput Sequencing
Methods such as MeRIP-Seq (methylated RNA immunoprecipitation followed by sequencing) and Pseudo-Seq (pseudouridine sequencing) allow for the genome-wide mapping of specific RNA modifications at single-nucleotide resolution. These techniques involve the immunoprecipitation of modified RNA fragments followed by high-throughput sequencing.
Reverse Transcription-Based Methods
Techniques like DART-Seq (deamination adjacent to RNA modification targets) and RiboMethSeq (ribosome methylation sequencing) rely on the differential reverse transcription of modified and unmodified RNA bases to identify modification sites. These methods provide an alternative approach to antibody-based techniques.
Functional Assays
To investigate the functional consequences of RNA modifications, researchers use various assays, such as ribosome profiling to assess translation efficiency, RNA stability assays to measure RNA decay rates, and RNA-protein interaction studies to identify modification-dependent binding partners.
Challenges and Future Perspectives
While epitranscriptomics has made significant progress in recent years, several challenges remain. One major challenge is the development of more sensitive and specific methods to detect and quantify RNA modifications, particularly those that are present in low abundance or in specific cell types or tissues.
Future research in epitranscriptomics will focus on elucidating the molecular mechanisms underlying the regulation and function of RNA modifications, as well as their role in human health and disease. The integration of epitranscriptomics with other omics technologies, such as proteomics and metabolomics, will provide a more comprehensive understanding of how RNA modifications influence cellular processes and organismal phenotypes.
Additionally, the potential therapeutic applications of epitranscriptomics will be explored, such as targeting RNA modification pathways for the treatment of diseases associated with dysregulated gene expression. The development of small molecule modulators of RNA-modifying enzymes and the use of RNA-based therapies that incorporate modified nucleotides are promising avenues for future research.
Further Reading
Cold Spring Harbor Perspectives in Biology, The Epitranscriptome in Translation Regulation
BioTechniques, Epitranscriptomics: Mapping Methods and Beyond
Trends in Pharmacological Sciences, The rise of epitranscriptomics: recent developments and future directions